Computational and
Dielectric Models
A. Different computational models
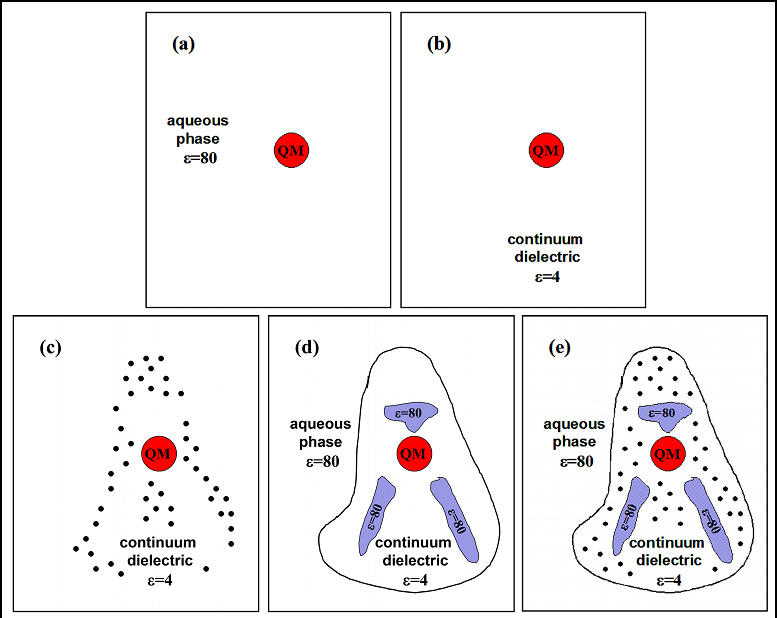
To get a sense how different structural details affect the apparent pKa values of His291 and Glu242, we considered several computational models. [Popovic, Quenneville, Stuchebrukhov, JPC B, 109, 3616 (2005); BBA 1757, 1035, (2006); Popovic & Stuchebrukhov, PPS, 5, 611 (2006)]
B. CuB center as QM-system (minimal model)
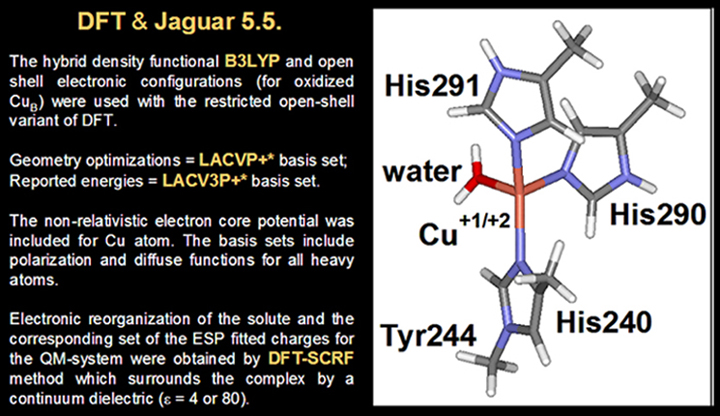
C. Binuclear complex as QM part of QM/MM (extended model)
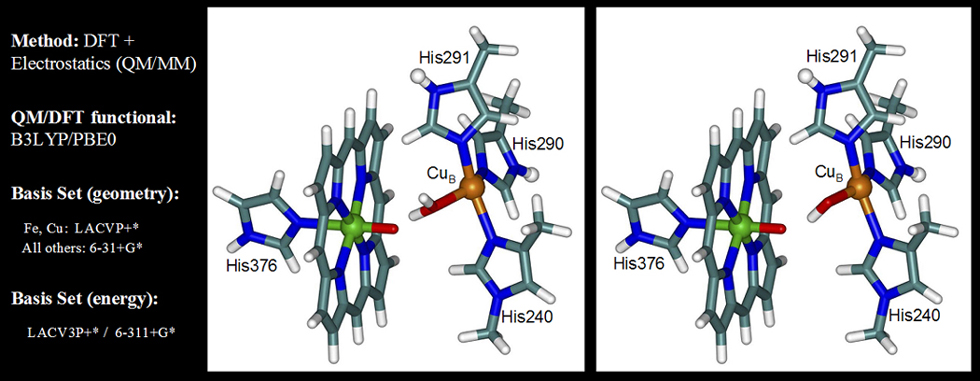
D. Geometry optimization specifics
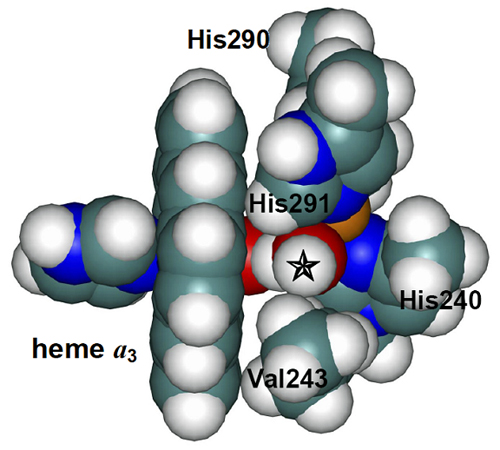
Space-filled rendering of the binuclear center using van der Waals radii. The figure shows how the Val243 residue affects the orientation of the H2O-CuB ligand. Without inclusion of the Val243 side chain, we were not able to obtain an appropriate geometry of the H2O/OH– ligand.
E. Full computational model
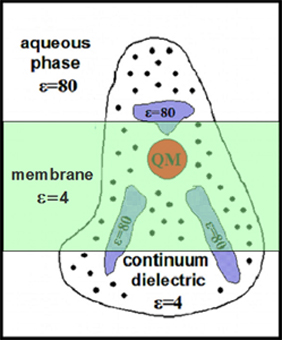
Final computations are done on a full computational model, for a minimal and extended complexes, for different redox states of the protein, and for the different dielectric constants assigned to the protein and water filled internal cavities.
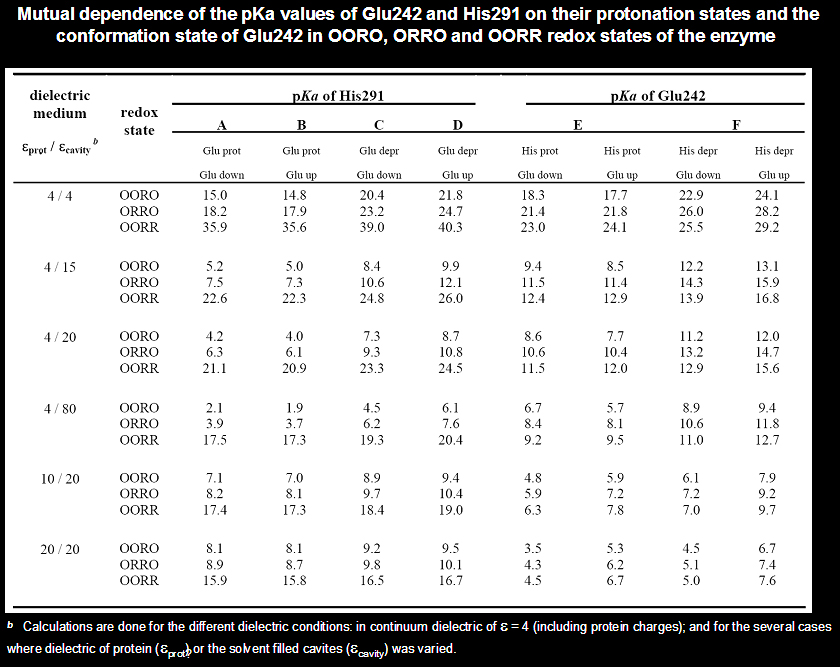
An example of recent calculations done for the different dielectric models of the protein-solvent system, diffrent conformations of Glu242 sidechain, and different redox states of CcO.