
results of prior research

Skin cancer is the most common form of cancer in the United States and can be lethal if not properly treated.
The leading cause of skin cancer is epidermal absorption of ultraviolet (UV) radiation.
DNA is believed to be the primary chromophore.
Indeed, photochemical reactions involving DNA have been linked to mutagenesis, carcinogenesis and cell death.
Fortunately, nature has designed a very elegant way of directly repairing damaged DNA for many organisms.
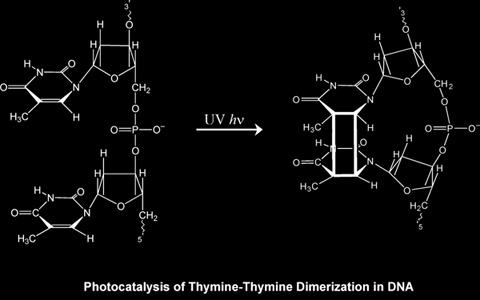
The most prevalent lesions formed by irradiation of DNA by UV-B or UV-C are the result of photoreactions of pyrimidine bases.
In particular, thymine dimers (T<>T) can form following photon absorption by a thymine monomer, intersystem crossing to its excited triplet state, and subsequent reaction with an adjacent thymine molecule.
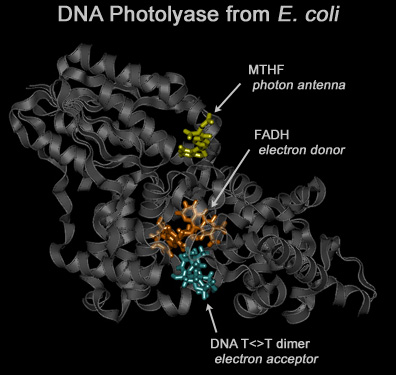 The photolyase enzyme reactivates DNA by direct repair of T<>T lesions.
It binds to the damaged DNA site, absorbs a (near UV-visible) photon, restores the pyrimidines to their monomeric forms, and dissociates from the substrate.
This repair mechanism is found in all three kingdoms of life but not in humans.
The photolyase enzyme reactivates DNA by direct repair of T<>T lesions.
It binds to the damaged DNA site, absorbs a (near UV-visible) photon, restores the pyrimidines to their monomeric forms, and dissociates from the substrate.
This repair mechanism is found in all three kingdoms of life but not in humans.
Our group is involved in theoretical and computational studies of the photo-initiated, electron transfer catalyzed DNA repair mechanism carried out by the photolyase enzyme.
The major effort in the past has been the study of the electron transfer reaction. (see Biophys. J., 76, 1241,
1999. and JACS
122. 1057, 2000.)
The work on DNA repair began as a side project on the application of electron transfer theories that we have developed.
The present thrust is toward an understanding of the molecular mechanism of photolyase and related issues of protein/DNA interactions.
Our research is supported by the NSF funded Center for Biophotonics, CBST, here on campus.
The structure of the photolyase enzyme has been solved some time ago(1DNP), however the structure of the DNA/Photolyase complex remained unknown until the end of 2004 (see below).
The topics we would like to address are:
1.) The mechanism of recognition of the dimers of DNA by photolyase. Our previous work demonstrated that this is a very inticate question since the dimer is not 'flipped out' of the DNA structure, but instead is 'poorly visible' on the surface of DNA unless photolyase is attached to it.
2.) The mechanism of sliding/diffusion of photolyase along DNA. What has also been suggested is that DNA itself may be processed through a fixed structure in the cell.
3.) In our previous work we showed that upon binding of photolyase to a dimer on DNA, the latter is flipped out of the DNA helix, and gets positioned deep in the catalytic site of photolyase, close to the key redox co-factor FADH.
In the repair reaction, this co-factor, upon transfer of photoexcitation, is electronically excited, transfers an electron to the dimer, the latter splits, and an electron is then returned back to FADH.
The mechanism by which photolyase flips the dimer needs to be clarified.
4.) Details of the repair reaction, which leads to the splitting of the dimer.
The figure below shows how the enzyme binds to DNA.
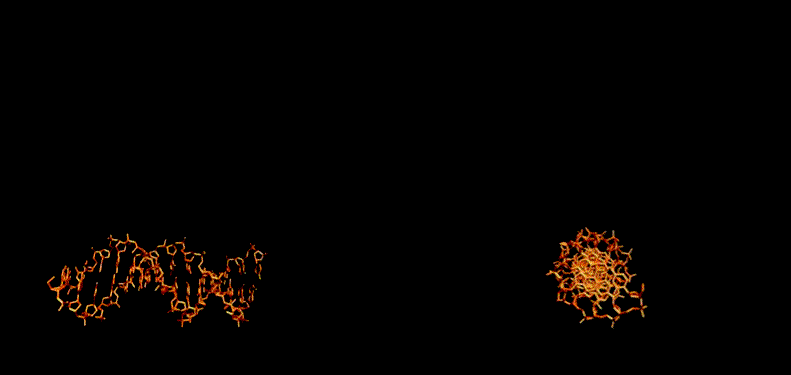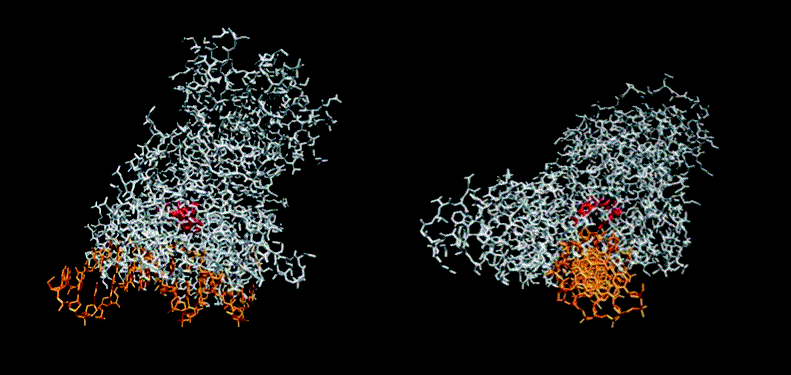
December 2004 - Incredible news!!! The stucture of DNA/Photolyase complex is solved experimentally and reported in Science. In our work of 2000-2001 we predicted the stucture of the complex, our prediction turned out to be correct to the minor details. As we predicted, upon enzyme binding to the dimer, DNA locally "melts", and the dimer flips out of the DNA helix.